Servers & Tools
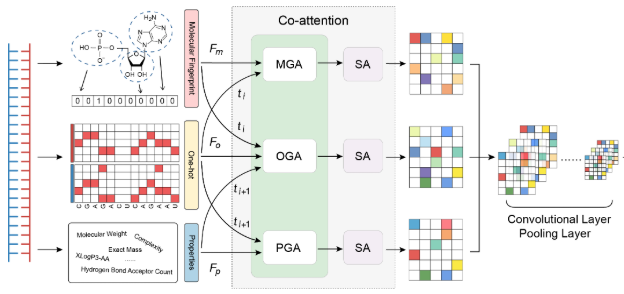
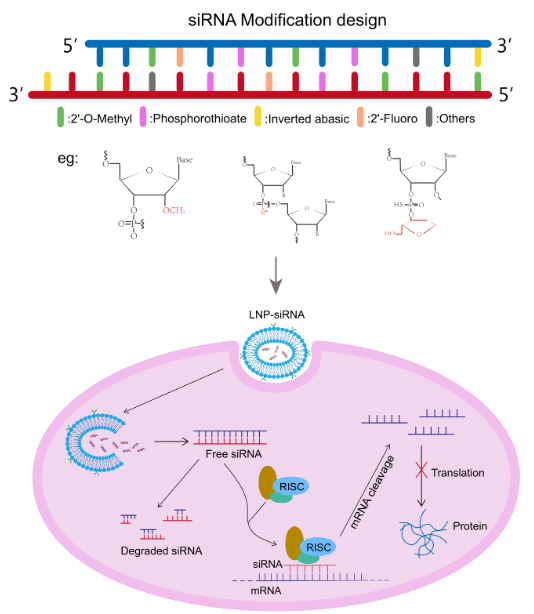
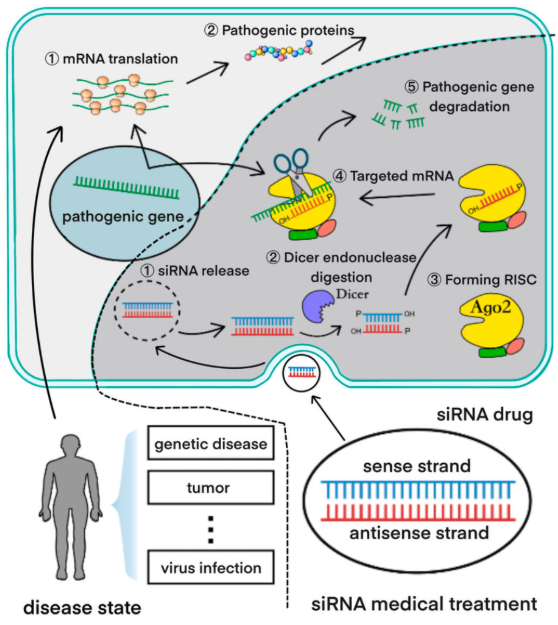
Cm-siRPredPredicting chemically modified siRNA efficiency based on multi-view learning strategy. |
|
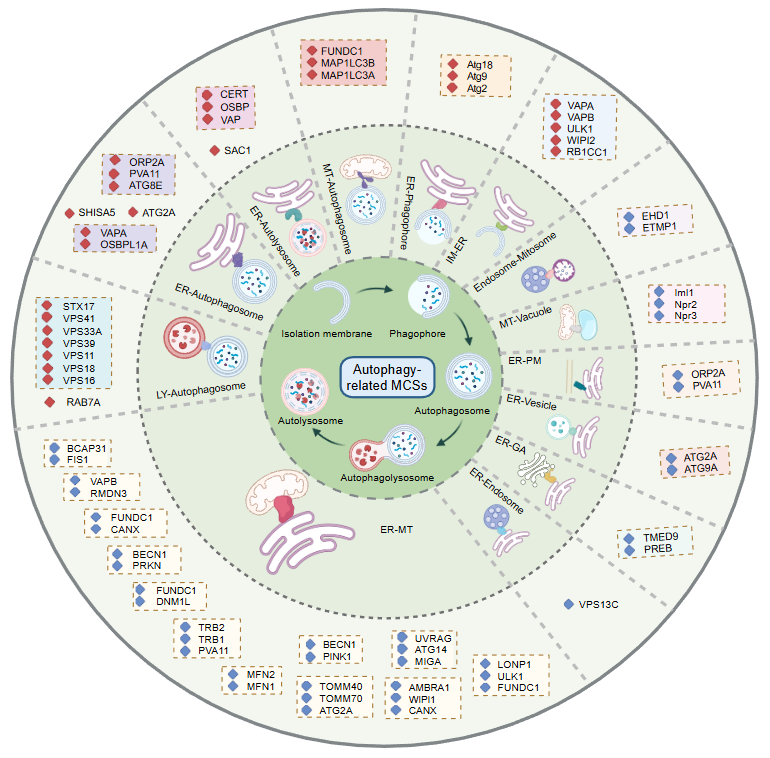
AutoMCS NavigatorMapping autophagy-related membrane contact site proteins and complexes. |
|
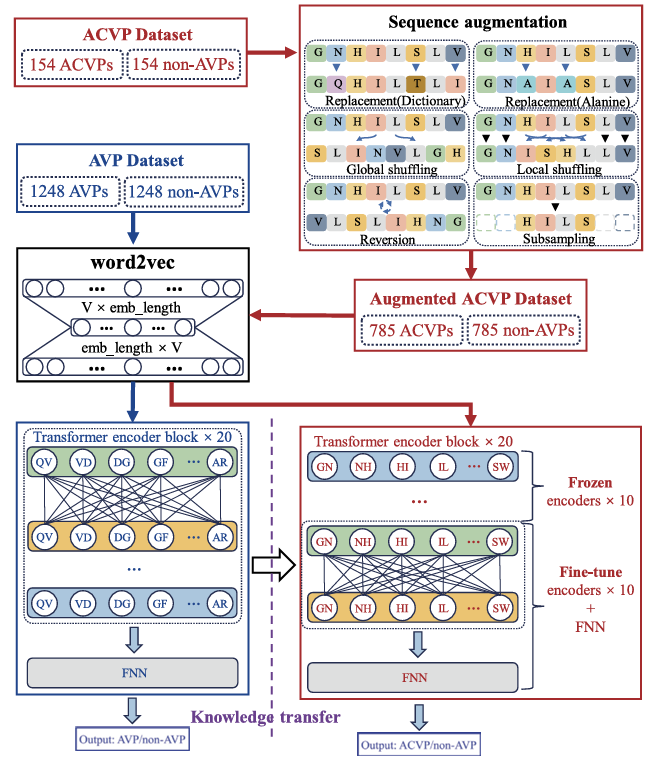
AcvpredEnhanced Prediction of Anti-Coronavirus Peptides by Transfer Learning Combined with Data Augmentation. |
|
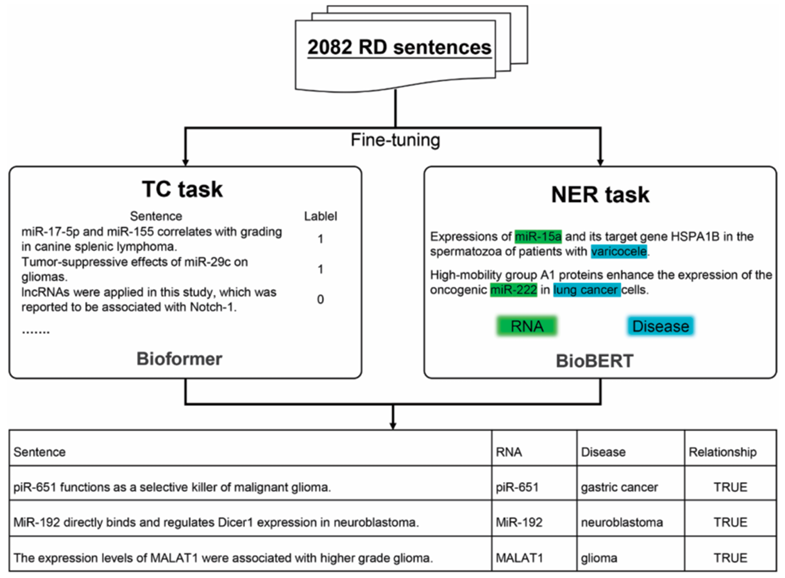
RDscanExtracting RNA-disease relationship from literature Using pre-training model. |
|
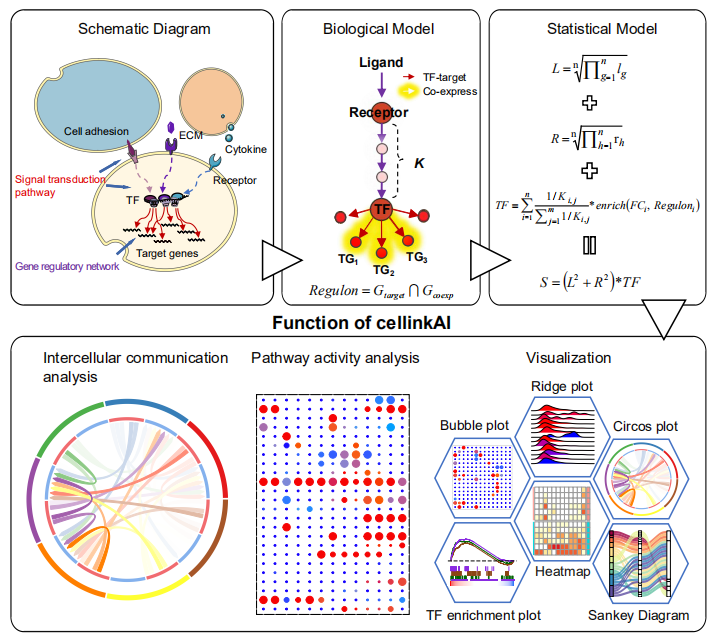
CellCallIntegrating paired ligand-receptor and transcription factor activities for cell-cell communication analysis. |
|
RIscoperA tool for extraction of RNA-RNA interactions from the literature. |
Databases
CMsiRNAdbA Database of Chemically Modified siRNA Silencing Efficiency for Nucleic Acid Drug Design. |
|
CellResDBDeciphering cancer therapy resistance via patient-level single-cell transcriptomics. |
|
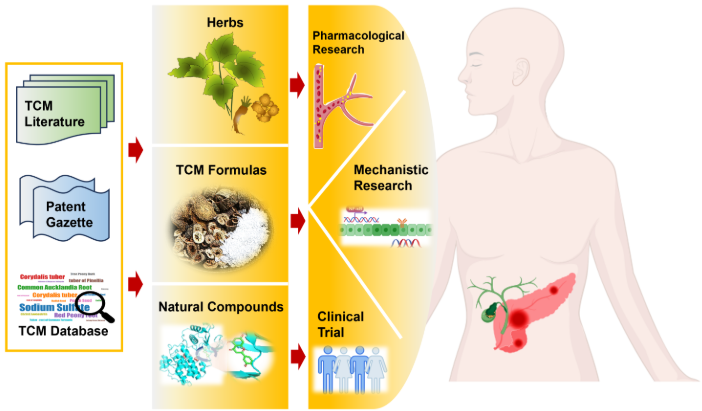
TCMAPUnderstanding the treatment of acute pancreatitis and its complications -a database for assessing traditional Chinese medicine use. |
|
siRNAEfficacyDBAn experimentally supported small interfering RNA efficacy database. |
|
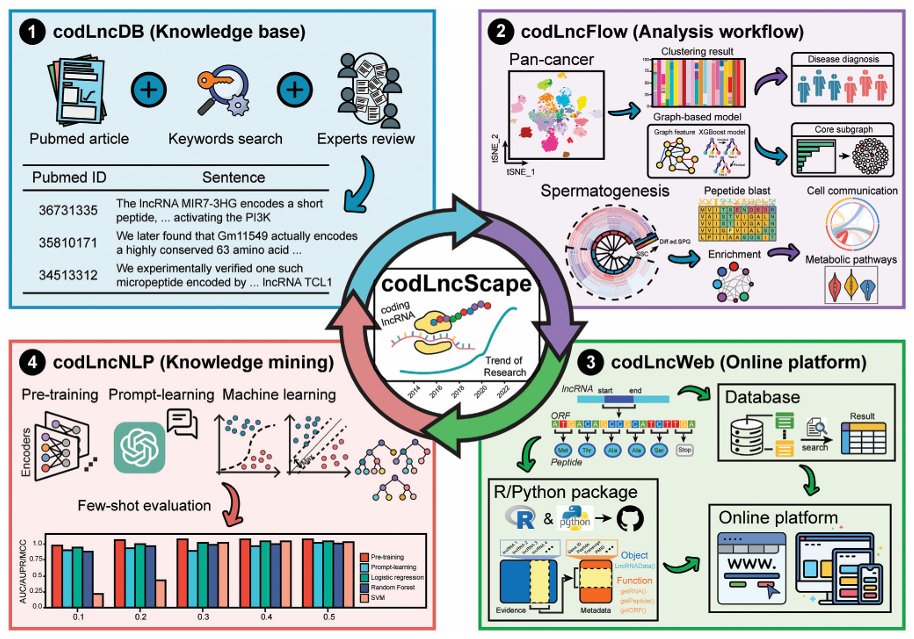
CodLncScapeA self-enriching framework for systematic collection and exploration of Coding LncRNAs. |
|
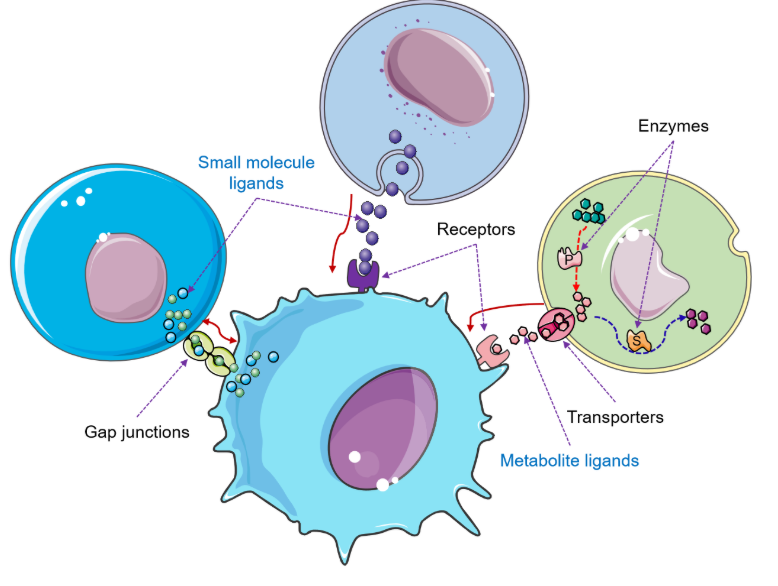
MRCLinkdbPredicting intercellular communication based on metabolite-related ligand-receptor interactions. |
|
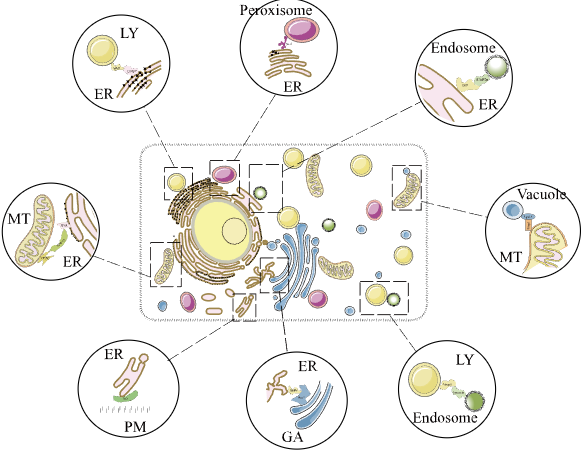
MCSdbA comprehensive database of proteins residing in membrane contact sites. |
|
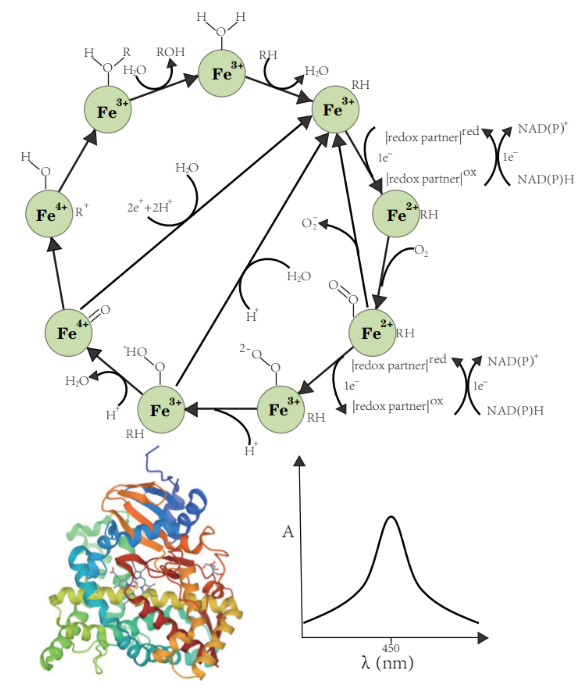
P450RdbA manually curated database of reactions catalyzed by cytochrome P450 enzymes. |
|
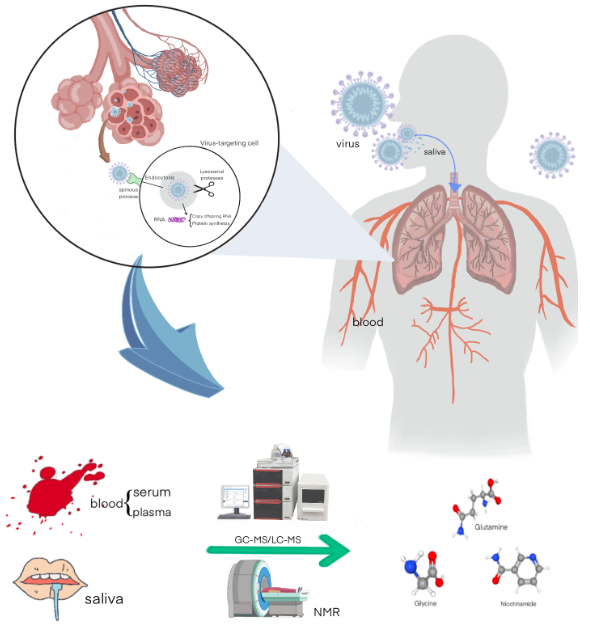
MetaboliteCOVIDA manually curated database of metabolite markers for COVID-19. |
|
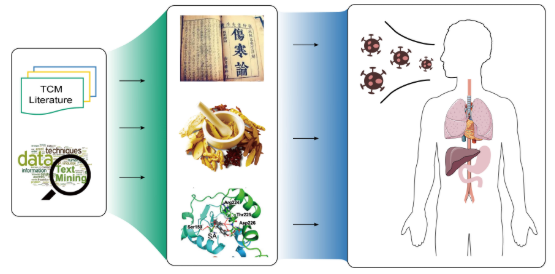
TCM2COVIDA resource of anti-COVID-19 traditional Chinese medicine with effects and mechanisms. |
|
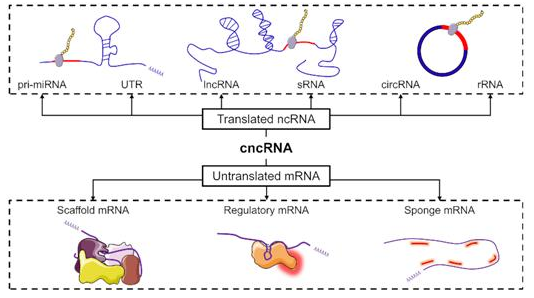
cncRNAdbA manually curated resource of experimentally supported RNAs with both protein-coding and noncoding function. |
|
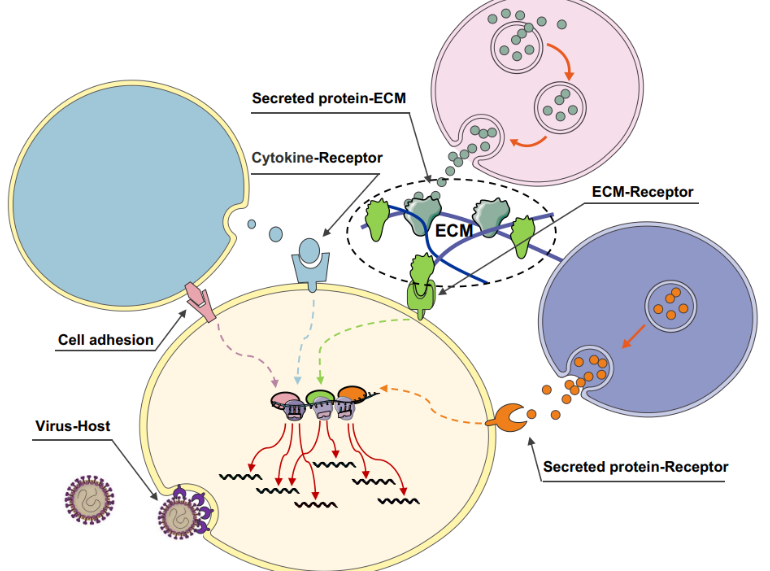
CellinkerA platform of ligand-receptor interactions for intercellular communication analysis. |